Reverse engineering morphogenesis through Bayesian optimization of physics-based models
Nilay Kumar1,
Mayesha Mim2,
Alexander Dowling2,
Jeremiah Zartman2
1. Purdue University
2. University of Notre Dame
01/23/2025
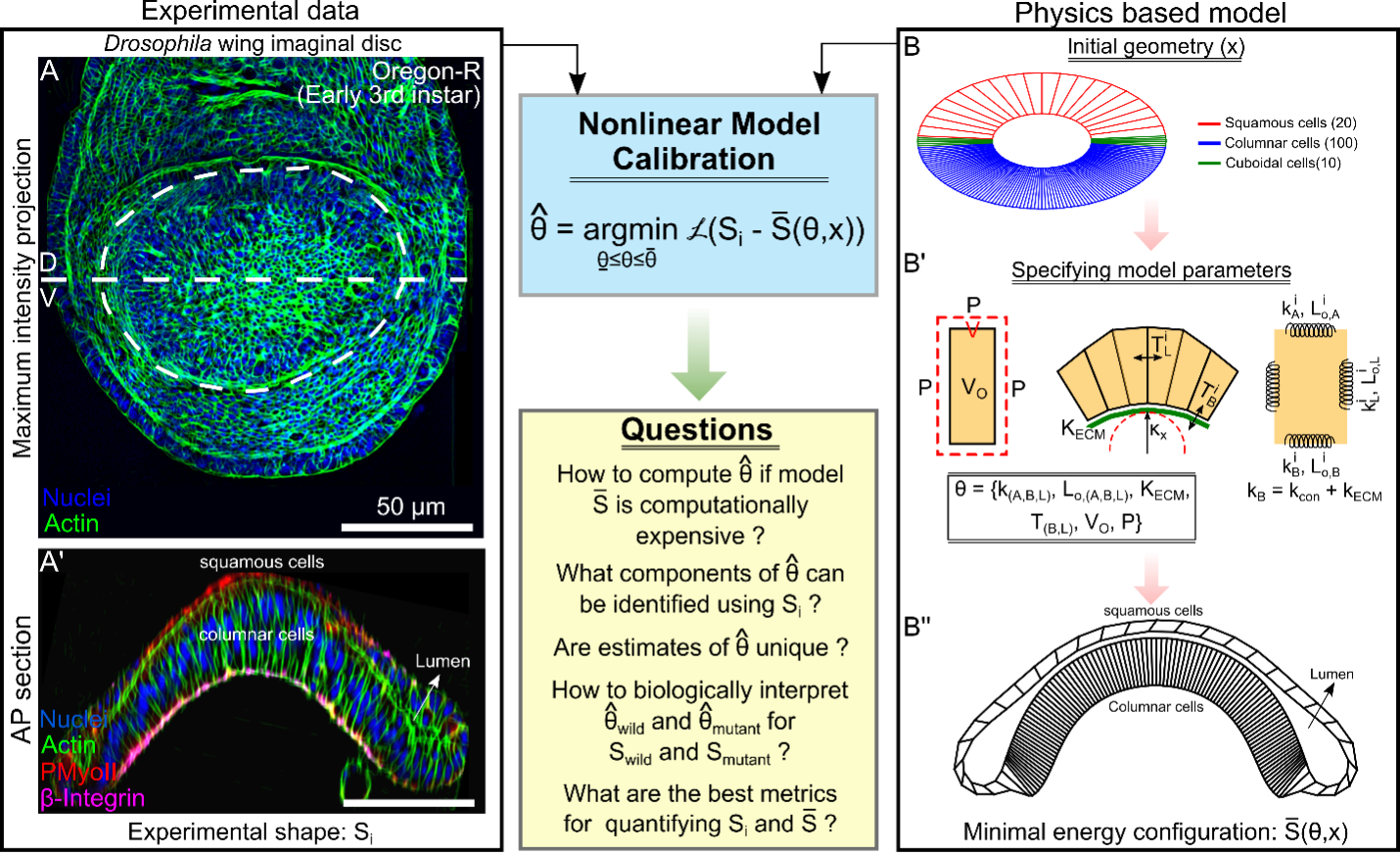
Morphogenetic programs coordinate cell signaling and mechanical interactions to shape organs. In systems and synthetic biology, a key challenge is determining optimal cellular interactions for predicting organ shape, size, and function. Physics-based models defining the subcellular force distribution facilitate this, but it is challenging to calibrate parameters in these models from data. To solve this inverse problem, we created a Bayesian optimization framework to determine the optimal cellular force distribution such that the predicted organ shapes match the experimentally observed organ shapes. This integrative framework employs Gaussian Process Regression, a non-parametric kernel-based probabilistic machine learning modeling paradigm, to learn the mapping functions relating to the morphogenetic programs that maintain the final organ shape. We calibrated and tested the method on Drosophila wing imaginal discs to study mechanisms that regulate epithelial processes ranging from development to cancer. The parameter estimation framework successfully infers the underlying changes in core parameters needed to match simulation data with imaging data of wing discs perturbed with collagenase.
Keywords
Bayesian optimization morphogenesis mechanotransduction extracellular matrixCite this dataset
Researchers should cite this dataset as follows:
Kumar, N., Mim, M., Dowling, A., Zartman, J. (2025). Reverse engineering morphogenesis through Bayesian optimization of physics-based models. Purdue Unversity EMBRIO Data Inventory.
License
The dataset is licensed under Attribution 4.0 International (CC BY 4.0).